
講演者・サマリー
When RNA is Not the Message: Ribozymes, Telomerase, and Long Noncoding RNAs Thomas R. Cech(コロラド大学ボールダー校教授)

In all of life on Earth, information flows from DNA to RNA to Protein. Thus, RNA has a role as a “message,” transmitting the information encoded in our chromosomal DNA. More recently, the biological importance of noncoding RNA has been revealed. RNA molecules can have active roles in biology, even acting as enzymes. Prof. Cech will describe the events that led to the discovery of the first catalytic RNA, or ribozyme, and subsequent work that revealed the structures and functions of these remarkable molecules. The finding that RNA could be a biocatalyst fueled speculation about a primordial RNA World, where RNA served as both information (genotype) and function (phenotype). Dr. Cech will then describe recent work on the structure and function of telomerase, the RNP (ribonucleoprotein) enzyme that synthesizes new DNA at chromosome ends. Finally, he will mention recent work on proteins that bind long noncoding RNAs (lncRNAs) in the human cell nucleus to regulate gene expression.
The Role of RNA and Proteins in the Removal of lntrons from Pre-mRNA John Abelson(カリフォルニア大学サンフランシスコ校・カリフォルニア工科大学名誉教授)

Eukaryotic genes contain introns, segments of DNA that interrupt the continuity of genetic information. In gene expression the introns are removed from an RNA transcript of the gene, the pre-mRNA. The reaction is a two-step trans-esterification process. In the first step of splicing the 2'OH of the conserved branch point adenosine attacks the phosphodiester bond at the 5' splice site, generating the 5’ exon and lariat-3’ exon intermediates. In the second step of splicing, the 3’OH of exon 1 attacks the phosphodiester bond at the 3’ splice site, forming the mRNA and lariat intron products.
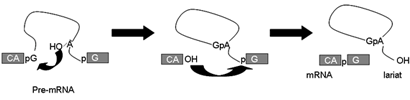
This reaction can proceed in vitro in a cell free extract and requires the addition of ATP. We now know that each splicing event requires the de novo assembly of a large supra-molecular complex, the spliceosome. The spliceosome contains five RNA molecules (snRNAs) in addition to the pre-mRNA and more than 100 proteins.
Quite remarkably it was discovered quite early that the Group II self-splicing RNAs remove the intron by the same two-step trans-esterification process shown above. This reaction does not require ATP or extrinsic proteins.
I will review evidence that the requirement for ATP in eukaryotic splicing is necessary for proofreading. Pre-mRNA splicing must be accurate to the nucleotide and errors result in non-functional mRNA products.
It seems likely that pre-mRNA splicing evolved from Group II transposons. High resolution structures of the Group II RNA have provided new insights into the mechanism of splicing.
The Nucleosome: Guardian and Gateway to the Genome Timothy J. Richmond (チューリヒ工科大学教授)

Chromatin is the substrate engaged by the molecular machines responsible for replication, recombination and transcription of the DNA genomes of eukaryotic organisms. The nucleosome is the fundamental repeating unit of chromatin, and nucleosome mapping in situ has shown that nucleosome positions along DNA determine access to the regulatory sequences essential to these nuclear processes. Nucleosome position is determined chiefly by DNA sequence and by ATPdependent, chromatin remodeling factors. However, only limited relevant DNA structural information essential for a detailed understanding of nucleosome positioning is available. We have recently determined the X‐ray structure of a nucleosome from mouse mammary tumor virus (147 bpMMTV‐A) which reveals new sequence‐dependent DNA conformations. This crystal structure suggests a method for obtaining high resolution structures for many different DNA sequences bound in a nucleosome. We have also determined the X‐ray structure of the chromatin remodeling factor ISW1a(ΔATPase) from S.cerevisiae bound with DNA, and imaged it in nucleosome complexes using cryo‐electron microscopy. We show how this chromatin remodeling factor, which acts as a genespecific transcription repressor, can set the spacing between two adjacent nucleosomes measuring the distance between them. Structural studies of ISW1a are progressing using both mono and dinucleosomes.

